Kyle's Blog
Building a 3D Map in R — Covid Density in the US by Income
Oct, 2020
We are in a golden age of data visualizations AND data!
First, lots of libraries
# Lots of libraries
library(sp)
library(choroplethrMaps)
library(tidyverse)
library(tidyverse)
library(rgl)
library(rayshader)
library(png)
library(ggplot2)
library(grid)
library(mapproj)
library(av)
Load and preprocess 1st data set
# Preprocess 1st data set
income = read.csv('income.csv', stringsAsFactors = F)
income2017 <- income[-c(3:36)]
income2017$X2017 = as.integer(gsub(',', '', income2017$X2017))
colNames = c('region', 'value')
colnames(income2017) <- colNames
map_outline = data(state.map)
income2017$region = tolower(income2017$region)
income2017 <- income2017 %>% mutate(., region =
replace(region, region=='d.c.', 'district of columbia') )
joinedData <- left_join(state.map, income2017, by = 'region')
keep = c(1,2,3,6,10,13)
joinedData <- joinedData[, keep]
map_data = joinedData
map_data$value = as.numeric(map_data$value)
map_data <- map_data %>% arrange(., order)
map_data <- map_data %>% filter(., region != 'alaska')
map_data <- map_data %>% filter(., region != 'hawaii')Map
# Map
map_us = ggplot(map_data, aes(long, lat, group=group, fill =
value)) +
geom_polygon() + # Shape
scale_fill_gradient(limits=range(map_data$value),
low="#FFF3B0", high="#E09F3E") +
layer(geom="path", stat="identity", position="identity",
mapping=aes(x=long,
y=lat, group=group,
color=I('#FFFFFF'))) +
theme(legend.position = "none",
axis.line=element_blank(),
axis.text.x=element_blank(), axis.title.x=element_blank(),
axis.text.y=element_blank(), axis.title.y=element_blank(),
axis.ticks=element_blank(),
panel.background =
element_blank()) +
coord_map(projection =
"mercator")
map_us
# Save as PNG
xlim = ggplot_build(map_us)$layout$panel_scales_x[[1]]$range$range
ylim = ggplot_build(map_us)$layout$panel_scales_y[[1]]$range$range
ggsave('map_us.png')Load and map 2nd data set
# Load and map 2nd data set
covid_df = read.csv('covidbycounty.csv')
geocode_df = read.csv('geocodedcounties.csv')
uniqueGeocode <- geocode_df[,-1]
uniqueCountyGeocode <- unique(uniqueGeocode)
uniqueCountyGeocode$state_county <-
paste0(uniqueCountyGeocode$state, uniqueCountyGeocode$county)
uniqueCountyGeo$county <- paste0(uniqueCountyGeo$county, "
County")
uniqueCountyGeo <-
uniqueCountyGeo[!duplicated(uniqueCountyGeocode$state_county), ]
covid_df = left_join(covid_df, uniqueCountyGeo[c(2,3,4,5)], by
=c('county', 'state'))
covid_df = na.omit(covid_df)
covid_df <- covid_df %>% filter(., state != 'HI')
covid_df <- covid_df %>% filter(., state != 'AK')
map_us_png = readPNG('map_us.png')
us_covid_deaths = ggplot(covid_df) +
annotation_custom(rasterGrob(map_us_png,
width=unit(1,"npc"), height=unit(1,"npc")),
-Inf, Inf, -Inf, Inf) + # Background
xlim(xlim[1]-2,xlim[2]+1) + # x-axis Mapping
ylim(ylim[1]-6,ylim[2]+6.5) + # y-axis Mapping
geom_point(aes(x=longitude, y=latitude, color=deaths),
size=0.1) +
scale_colour_gradient(name = 'deaths',
limits=range(covid_df$deaths),
low="#FCB9B2", high="#B23A48") +
theme(axis.line=element_blank(),
axis.text.x=element_blank(), axis.title.x=element_blank(),
axis.text.y=element_blank(), axis.title.y=element_blank(),
axis.ticks=element_blank(),
panel.background =
element_blank())
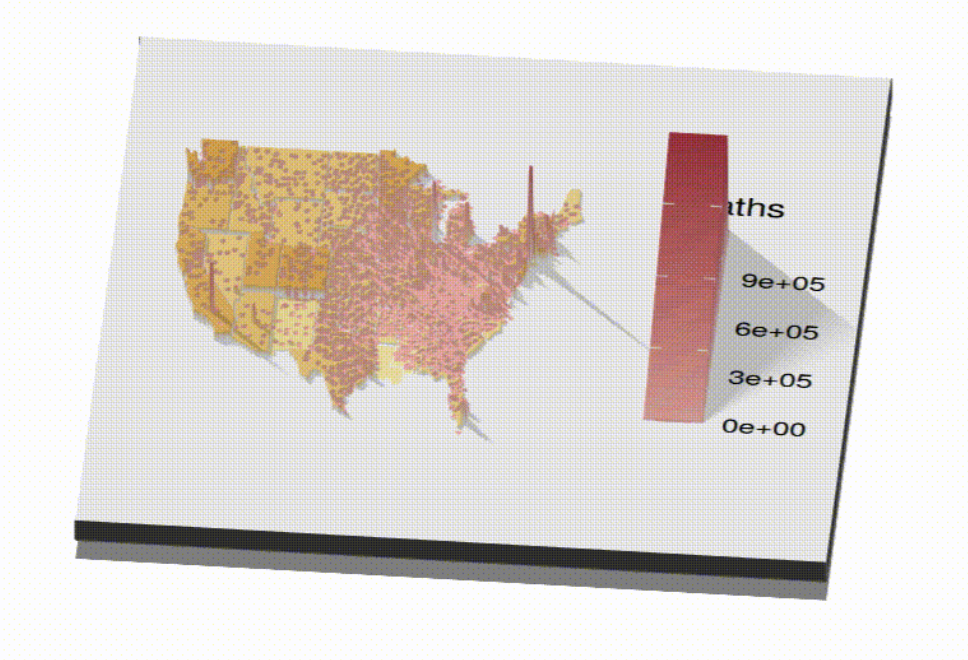
us_covid_deaths
ggsave('covid_deaths.png')Make the points 3D and record video
# Make the points 3D and record video
plot_gg(us_covid_deaths, multicore = TRUE)
par3d(windowRect = c(0, 0, diff(xlim) * 2500, diff(ylim) * 2500))
render_camera(fov = 70, zoom = 0.2, theta = 30, phi = 20)
render_depth(focus = 0.8, focallength = 600)
phivechalf = 30 + 60 * 1/(1 + exp(seq(-7, 20, length.out =
180)/2))
phivecfull = c(phivechalf, rev(phivechalf))
thetavec = 0 + 45 * sin(seq(0,359,length.out = 360) * pi/180)
zoomvec = 0.45 + 0.2 * 1/(1 + exp(seq(-5, 20, length.out = 180)))
zoomvecfull = c(zoomvec, rev(zoomvec))
render_movie(filename = 'output1', type = "custom", frames =
360, phi = phivecfull, zoom = zoomvecfull, theta = thetavec)
transition_values <- function(from, to, steps = 10, one_way =
FALSE, type = "cos") {
if (!(type %in% c("cos", "lin"))){stop("type must be one
of: 'cos', 'lin'")}
range <- c(from, to)
middle <- mean(range)
half_width <- diff(range)/2
if (type == "cos") {scaling <- cos(seq(0, 2*pi /
ifelse(one_way, 2, 1), length.out = steps))}
else if (type == "lin"){
if (one_way) {xout <- seq(1, -1, length.out
= steps)}
else {xout <- c(seq(1, -1, length.out =
floor(steps/2)), seq(-1, 1, length.out = ceiling(steps/2)))}
scaling <- approx(x = c(-1, 1), y = c(-1,
1), xout = xout)$y }
middle - half_width * scaling
}
theta <- transition_values(from = 0, to = 360, steps = 360,
one_way = TRUE, type = "lin")
phi <- transition_values(from = 10, to = 70, steps = 360,
one_way = FALSE, type = "cos")
zoom <- transition_values(from = 0.4, to = 0.8, steps = 360,
one_way = FALSE, type = "cos")
render_movie(filename = 'output2', type = "custom", frames =
360, phi = phi, zoom = zoom, theta = theta)
rgl.close()